
You can also view the sequences aligned in pairwise mode, along with a matrix displaying the identities and similarities of each pair of sequences.Ĭopyright © 2022 MacVector, Inc. MacVector: sequence comparisons using a matrix method Methods Mol Biol. Bioz Stars score: 86/100, based on 1 PubMed citations. One window provides a high quality customisable graphical text display designed for output on a laser printer, while another duplicates the output from ClustalW. MacVector: sequence comparisons using a matrix method. Clustalw Protein Alignment, supplied by MacVector, used in various techniques. The aligned sequences can be displayed in result windows in a variety of different ways. MacVector provides a variety of algorithms that you can use to analyze the composition of protein sequences. For example, you can set up the editor so that all matches to the consensus are displayed as dots - this is very useful for identifying variant regions in otherwise closely related sequences. MacVector will also retrieve protein or nucleic acid sequences of interest from databases by conducting an Internet search, which is trivial to set up as. There are a variety of parameters and modes that control how the residues appear on-screen. Contiguous sequence was constructed using MacVector ® Assembler. Sequences can be viewed using plain monochrome text or colored according to user-defined color groups. DNA sequences were analyzed and protein predictions made using MacVector ® v10.6 (MacVector Inc, Cary, NC). You work with alignments in the multiple sequence alignment editor. MacVector supports the following multiple sequence file formats: You can also directly open an existing file containing multiple sequences. Clustal Sequence Alignment, supplied by MacVector, used in various techniques. You can create alignments from sequence already open in MacVector, or by creating an empty alignment and populating it with sequences imported from disk. pairs of DNA sequences, but also a DNA sequence with a protein sequence. The MacVector single sequence Editor will show those (click and hold on the Display toolbar button) but if you select and copy, only the DNA sequence (with any overlapping features) will be copied to the clipboard. The resulting primer sequences were further analyzed for consensus alignment in silico against reference sequences grouped by clade membership. The alignments can be output in a variety of formats or used to generate phylogenetic reconstructions using a number of built-in algorithms. MacVector can search DNA sequences for restriction sites using any or all of. All sequence alignments, mismatch identification, and analyses of temperature characteristics were made in silico using tools in MacVector software (v. programs (for example PC/GENE, Lasergene, MacVector, Accelrys etc.). MacVector uses the ClustalW algorithm to automatically align any number of sequences and provides a sophisticated editor that lets you fine tune the alignments. Analysis of nucleotide and protein sequence data was initially restricted to those. You can use MacVector to align related DNA or Protein sequences. Alternatively, there is ample freeware or. Many available sequence packages, like Vector NTI or MacVector, contain built-in protein analysis software that can predict potential phosphorylation sites. Full pricing for the application and its licensing conditions are available here.Sequence Analysis Tools for Molecular Biologists Thanks to the power of computing and ingenious programmers, you can use online programs to predict potential phosphorylation sites in your protein.

MacVector 10.6 requires Mac OS X 10.3.9 or later to install and run. Other features of the new version include enhancements and optimizations to the Align To Reference and Assembler alignment tools.
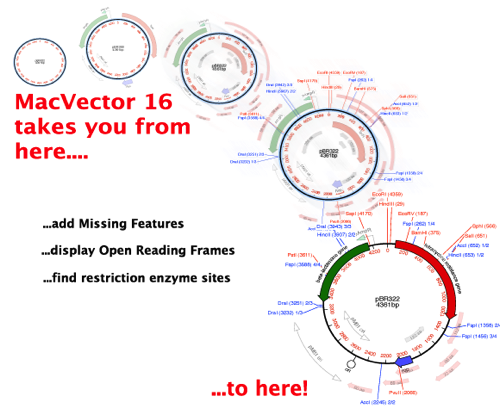
Creating dot plots of DNA to DNA, Protein to Protein and DNA to protein. It also has the added benefit of migrating any old and deprecated feature information contained in the Vector NTI file into the current nomenclature. MacVector is a commercial sequence analysis application for Apple Macintosh. The new version reads all of the standard features and annotations associated with each sequence, follows the Genbank format for the features table and is always kept up to date with the latest Genbank release. Version 10.6 includes several new features, including the introduction of a Vector NTI database import function that allows MacVector to directly read sequence data from databases created by Vector NTI Advance v10 or earlier, even if the database resides on a Windows machine accessible over a network. Software developer MacVector has released MacVector 10.6, the company's DNA and protein sequence analysis program. Locating gaps in amino acid sequences to optimize the homology between two proteins.


 0 kommentar(er)
0 kommentar(er)
